Alzheimer’s is a devastating disease. Does it have to stay that way? A recent genomics study suggests it may not.
A 2019 study done by Kunkle et al. has associated five new genes with Alzheimer’s disease, this allows scientists to better understand how Alzheimer’s disease works, and to create medications that can help save and extend lives!
How scientists did this was that they took genes found in people with Alzheimer’s, and compared them to genes found in people without Alzheimer’s, a technique widely known as a Genome Wide Association Study (GWAS).
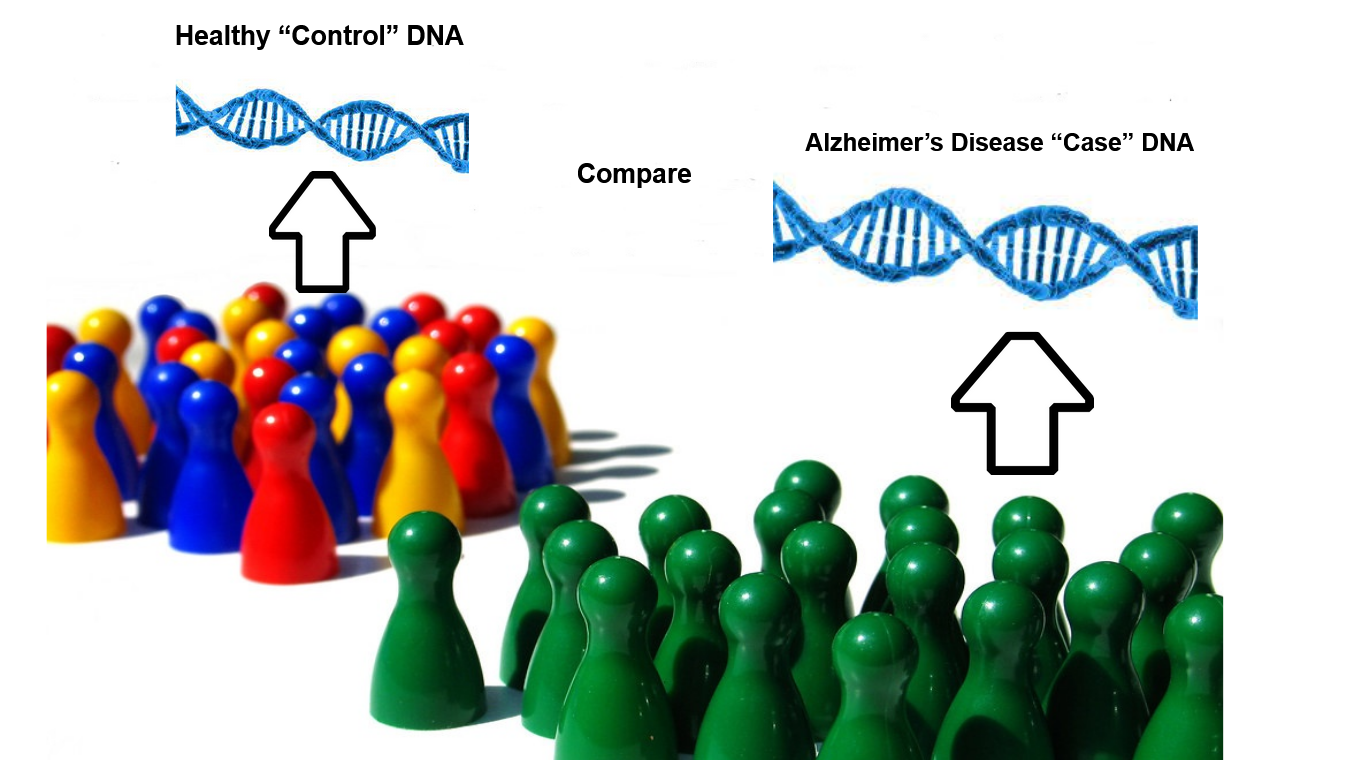
How a GWAS (genome-wide association study) is done. DNA is taken from people without Alzheimer’s disease, called “control” DNA, and compared to DNA from people with Alzheimer’s disease, called “case” DNA.
And what scientists can do with the information from a GWAS is that they can find out what gene variants are only present in people with a certain condition, in this case, Alzheimer’s disease, then study those variants to find out how they contribute to the ill state!
To better understand the genes that were uncovered through this study, it is necessary to get an overview of what Alzheimer’s disease is. To summarize, Alzheimer’s disease is a disorder where one’s brain begins to break down, in turn, affecting speech, memory, and thought. Two important features of Alzheimer’s disease are the presence of abnormal protein build up, also known as amyloid plaques, and twisted fibres, also known as tau tangles, in brain cells. These features are represented in the image below.
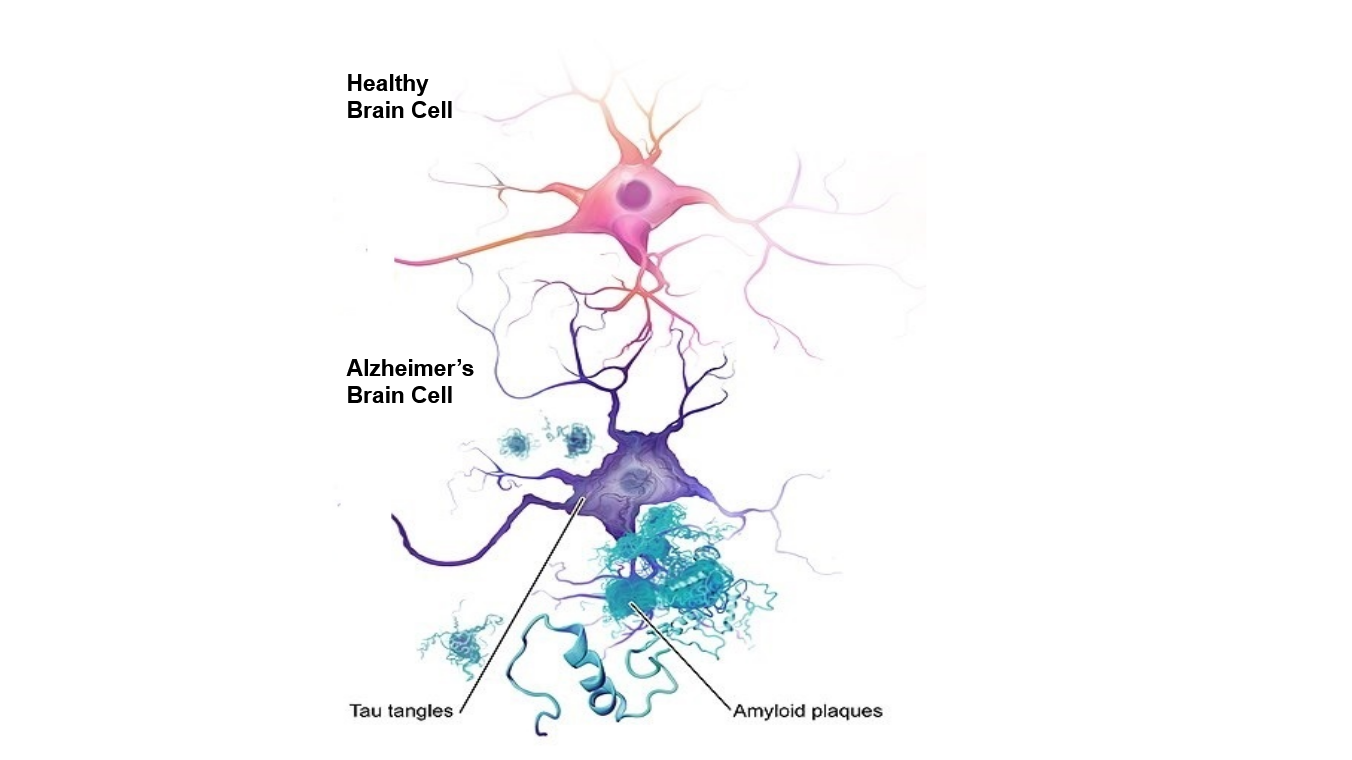
Healthy vs Alzheimer’s disease brain cell. Abnormal protein buildup (Amyloid plaques) and twisted fibres (Tau tangles) are key features of Alzheimer’s Disease.
Scientists are not completely sure how or why these features occur, but these discoveries have helped with diagnosis, and provided scientists with clues in order to help solve this disorder.
Alz.org states that currently, over 6 million people are diagnosed with Alzheimer’s disease in the US. However, in a disease such as this, it is not only those who are sick that are affected, but also their loved ones. Thus, the findings from GWAS studies are important in order to get closer to the reason why Alzheimer’s disease occurs.
Below is an example graph that researchers would look at in order to identify the important genes in a GWAS. It is formally known as a Manhattan plot, and essentially, any genes that are above the red dotted line, also known as the threshold, are potential genes that can lead to a disease.
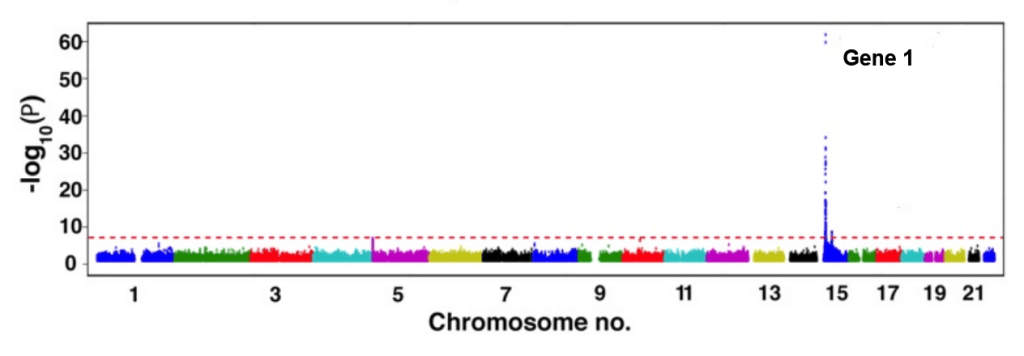
Example Manhattan plot. Any lines above the dotted red line, or threshold, depict a potential risk gene for a disease. In this case, Gene 1 would be a risk gene that scientists would identify through the plot and later study.
The genes identified by Kunkle et al. include: ADAM10, IQCK, WWOX, ACE, and ADAMTS1.
Sometimes, the genes found in GWAS do not provide information that is useful to understand a condition. A GWAS aims to uncover genes that tend to appear with a condition (association), even if they do not lead to it (causation). However, in this case, the genes uncovered by Kunkle et al. almost all provide reliable information as to what could cause Alzheimer’s disease, and what could help slow it down.
The image below depicts the connection between each uncovered gene to Alzheimer’s disease, and the studies that found these effects in brackets. A link to the studies can be found in the Reference section.
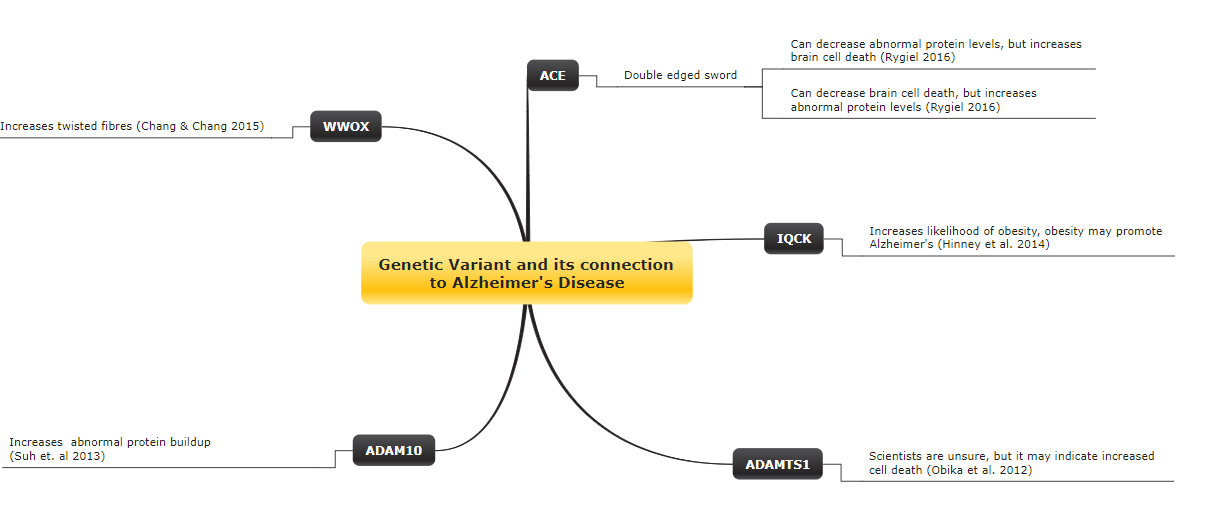
Concept map showing the connection between the identified gene(s) from Kunkle et al. to Alzhemier’s disease. Other studies that helped discover the effect(s) of a given gene are stated in brackets, and are linked at the end of the post.
As you can see from the concept map, Alzheimer’s disease is very complex. However, the discovery of these connections has allowed healthcare professionals and families alike to find ways to mitigate it!
For instance, on the healthcare professional end, this includes the production and authorization of drugs to help with the symptoms associated with Alzhemier’s disease. A newly approved method is the use of monoclonal antibodies, which essentially repurposes the body’s natural defense system to target and destroy the abnormal proteins that are building up.
On the family end, this includes dietary and lifestyle changes, which in the end, keeps everyone happy! Some examples of this include increased exercise and the consumption of foods high in omega-3, such as fish, which also reduces abnormal protein build up! As stated by Lim et al.
The study done by Kunkle et al. pays a lot more attention to detail than presented here. For instance, scientists could not just simply take the values found from the GWAS study and say that, because a given allele appears more in people with Alzheimer’s disease, it is therefore associated with it. They also had to ensure factors outside of the variables they measured did not impact their results, which are known as confounders. Scientists could not account for all the confounders, for many may be unknown, but the ones they did account for include: age, sex, and community.
These adjustments and a number of other things not only allowed the scientists to narrow down their findings to genes that had more potential in playing a role in Alzheimer’s disease, but also, led to two key findings.
One, that abnormal protein build up appears in both types of Alzheimer’s disease, which potentially allows them to use the same therapies for both.
And two, there’s a connection between the immune system and the mind, which allows scientists to potentially target the mechanisms that connect these two domains in order to better mitigate Alzheimer’s disease and other mind-related disorders. Which is related to the monoclonal antibodies previously mentioned.
These findings and the resilience shown by the scientists ultimately displays the amount of work and effort people are putting into ensuring a brighter future!
Overall, it is shown that GWAS yield fruitful results and high promise when attempting to understand certain conditions, such as Alzheimer’s disease. Thus, society has a lot to hope for in regard to health, disease, and well-being.
Learn more about Alzheimer’s disease
- What’s it like having Alzheimer’s disease?
- Alzheimer’s vs Dementia
- Aduhelm, a monoclonal antibody for Alzheimer’s disease
References
- Kunkle, B., Grenier-Bolet, B., et al. (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Retrieved April 05, 2021 https://www.nature.com/articles/s41588-019-0358-2#Abs1
- Chang, J., & Chang, N. (2015). WWOX dysfunction INDUCES sequential aggregation of Trappc6aδ, TIAF1, tau and amyloid Β, and causes apoptosis. Retrieved April 05, 2021, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4981022/
- Hinney, A., Albarayk, O., Antel, J., Volckmar, A., & Hebebrand, J. (2014). Genetic variation at the CELF1 (CUGBP, elav-like family Member 1 Gene) Locus is genome-wide associated with Alzheimer’s disease and obesity. Retrieved April 05, 2021, from https://pubmed.ncbi.nlm.nih.gov/24788522/
- Obika, M., Ogawa, H., et al. (2012). Tumor growth inhibitory effect of adamts1 is accompanied by the inhibition of tumor angiogenesis. Retrieved April 05, 2021, from https://onlinelibrary.wiley.com/doi/full/10.1111/j.1349-7006.2012.02381.x
- Rygiel, K. (2016). Can angiotensin-converting enzyme inhibitors impact cognitive decline in early stages of Alzheimer’s disease? An overview of research evidence in the elderly patient population. Retrieved April 05, 2021, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5105210/
- Suh, J., Choi, S., Romano, D., Gannon, M., Lesinski, A., Kim, D., & Tanzi, R. (2013, September 19). ADAM10 missense MUTATIONS Potentiate β-Amyloid accumulation by impairing prodomain Chaperone Function. Retrieved April 05, 2021, from https://www.sciencedirect.com/science/article/pii/S0896627313007940
About the author
This post was written by Kyle Valentino. He is entering his fourth year at the University of Toronto Scarborough. His plans for graduate school are undecided, but he has a strong interest in genetics and genomics. Among other things, Kyle likes to motivate those around him to think critically and become the best version of themselves. Thus, Kyle likes to spend time reading literature, and interacting with his peers to understand what can best help them reach their current goals. When Kyle is not doing this, you can find him listening to his favourite music, playing basketball, watching anime, or with family. You can contact Kyle at kyle.valentino@mail.utoronto.ca.

Open for questions!
LikeLike